eProtein Discovery™ Instrument and Software
Release notes
Introduction
This document describes the software updates and improvements to:
- eProtein Discovery Instrument Software
- eProtein Discovery Experiment Workflow versions
- eProtein Discovery software (Cloud-based)
Workflow versions can be identified when users start designing a new experiment. Release versions and description of change will be updated on this document periodically.
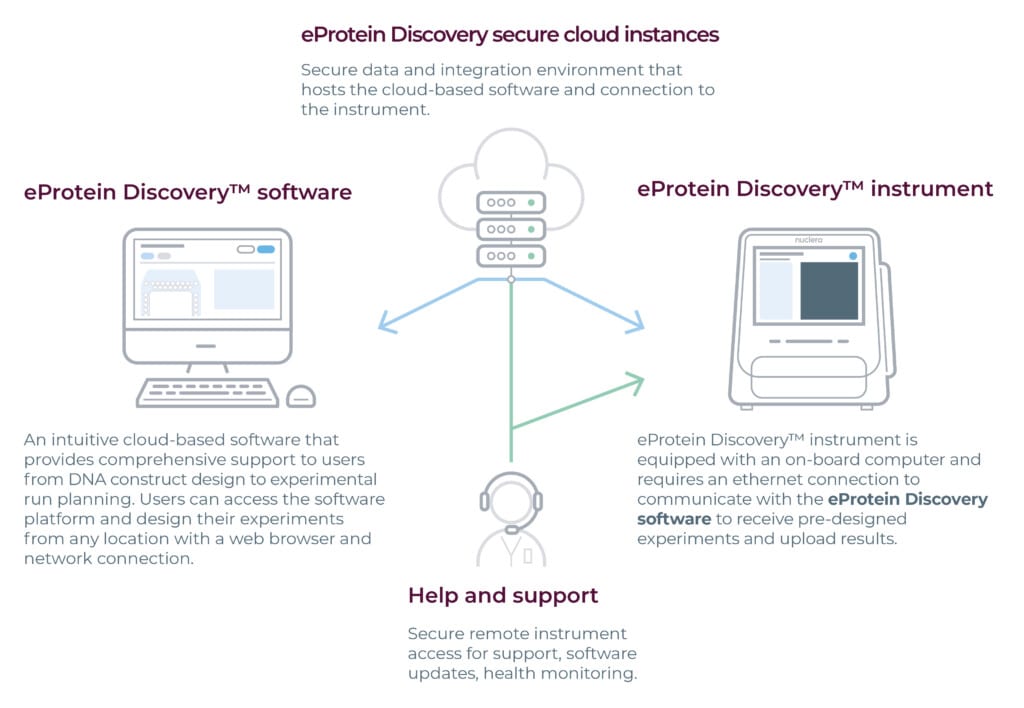
The above figure describes the eProtein Discovery™ software, instrument software and cloud-based architecture.
List of Releases:
New workflow release: eProtein Discovery™ Screen Experiment 5.2.2 workflow
⚠ This workflow will only be available on instruments that have been updated to software version 5.0.0. The following features will not be available retrospectively.
- Fix for cartridge pins not retracting when retrying a cartridge connection failure
- Fix for cartridge pins not retracting when starting a new experiment after a previous experiment was aborted or failed
- Scripts controlling droplet movements have been updated for more robust droplet control
eProtein Discovery™ Instrument software release 5.0.0
User interface updates:
- Instrument software updates are now automatically performed when the instrument is powered on, connected to the network, and idle. By default the updates occur at 1 am, but this time can be changed in the settings menu.
- The instrument time zone can now be adjusted on the settings screen
- Run scripts are added to each video that detail the actual events occurring during the workflow
- Instrument shutdown function is disabled if run report generation is in progress
- The "Open/Close Drawer" and "Connect/Disconnect Cartridge" options in the side menu are now disabled during experiments to prevent accidental cartridge detachment during a run
Bug fixes:
- The operating system has been updated to address a memory leak issue that could lead to system shutdowns.
- Fix for missing 3C Protease scissors icons and legend in PDF report
- Fix for occasional hang during PDF generation
- Fix for temperature control being left on after an experiment ends
- A correction has been implemented to resolve access issues for new users who were unable to log in to the instrument.
eProtein Discovery (cloud) software release
- eProtein Discovery Cloud software will be locked for 15 minutes after 10 successive unsuccessful sign-in attempts in a 24 hour period. This feature is designed to protect users from unauthorized access and enhance account security
- A link to the Nuclera Knowledge base is now accessible through the Help menu
- The experiment design page now supports the addition of up to 16 custom additives. Users can select ADD01-ADD16 in the additive selection panel and are encouraged to list the identify of each additives in the Notes section
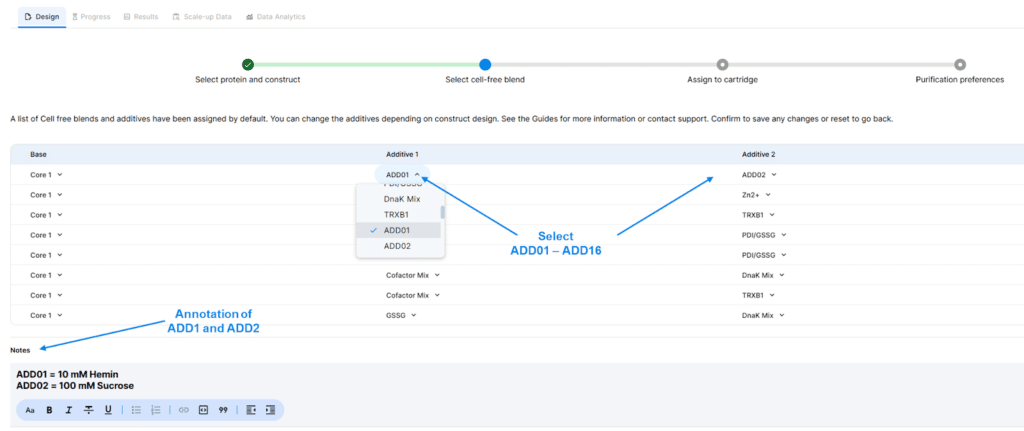
eProtein Discovery™ (cloud) software release
- Incompatible workflow experiment : With the automatic instrument software update in place, experiment workflow version may not be compatible with the instrument software. If that is the case, the cloud software will display a warning next to the workflow selection and a warning when the user attempts to finalize an experiment using a workflow experiment that is incompatible with one or more instruments within the user network.
- Support for import of fasta files compatible with sequence exported from Uniprot
Bug fixes and improvements:
- The protein sequence import page has been updated to provide better explanations if sequence incompatibilities are found
- Direct sequence input from Uniprot formatted fasta files is now enabled
- The eGene normalization csv file has been updated to include guidance for adding in very high or very low DNA concentrations
- Data analytics tab has been updated to correct information displayed in the pop up box when a cursor hovers over a data point on the lollipop chart
- Archived proteins are now excluded from file export
- DNA and protein sequences can now be exported with or without adapter sequences. The following file download formats are now supported:
- CSV
- FASTA

eProtein Explorer: Here's what's new in Version 1.3
- eProtein Explorer data analytics is now compatible with Firefox
The eProtein Discovery results file upload in eProtein Explorer is now compatible with the Firefox web browser, resolving an issue where valid report files were previously rejected for Firefox users.
New workflow release: eProtein Discovery™ Screen Experiment 5.1.0
⚠ This workflow will only be available on instruments that has been updated to software version 4.6.2. The following features will not be available retrospectively.
- A new “Data Analytics” tab has been added that display charts of the experiment results
eProtein Discovery™ (cloud) software release
- Users now have the capability to bulk import protein sequences from a fasta file by clicking on the + New Protein icon. This enhancement is particularly beneficial for users looking to import 24 sequences or more at once.
- New status icons have been introduced in the "protein tab," allowing users to quickly view and check run status of each protein without needing to navigate to individual protein page for details.
- Experiment can now be duplicated and renamed
- Improved validation of DNA synthesis complexity:
- Complexity score with IDT recommendation will be displayed in full
- A warning will be presented if DNA synthesis complexity is above a threshold of 10
- High complexity AA sequences are blocked from proceeding to the next step
- Protein sequences can now be exported in csv/fasta format
- Improved eGene normalization and sequence calculation in the “Possible Constructs” tab
- Addition of support links, bold, italics etc to notes field in proteins, projects and experiments
- Amino acid sequences for all possible constructs are now available in the “Possible Construct” table
- Protein sequences can now be exported in csv or fasta format by clicking on the ellipsis at the end of the protein list
- Several bug fixes to improve user experience
Instrument software release version 4.6.2
- Users can now export data to their SMBv3 network share on the local network directly from the instrument.
- USB exports have been improved to show export progress
- Experiments on online instruments are now automatically deleted after 30 days (only if all run data has been uploaded to the cloud), users can also manually delete experiments to free up disk space
- Various bug fixes and stability improvements
eProtein Discovery (cloud) software release
- Improvements to protein entry:
- DNA synthesis compatibility checks are more robust
- Several improvements to the “Results” and “Scale-up Data” tabs, which will be available for experiments created using the workflow “eProtein Discovery Screen Experiment 4.0.0”
- In the Expression Yield Result table in the Results tab, expression conditions that were selected for purification are indicated with a blue tick
- eGene recipes that include the 3C protease additive are now annotated with a scissor icon in measurement tables
- The Scale-up Data tab now features an interactive graph which allows users to visualize predicted yields of constructs which were screened and purified.
- Constructs and conditions for scale up can be added to the Scale-up table by selecting from the graph. Scale-up volume and predicted yield can be adjusted to meet desired yield. The data from this table generates a downloadable .csv with calculated volumes to facilitate scale up set up at the bench.
- Bug fixes:
- The “Possible constructs” tab is only visible after a protein is finalized
- The “Archive” button is re-positioned for improved visibility
- Any runs which were created with a protein that was later archived will have a warning when they get finalized
Instrument software release version 4.6.0
- AI/ML-based automated droplet quality checks enabled. This feature will improve droplet quality checks and flag any compromised reactions to the user
- Process improvements:
- PDF generation and report now downloadable straight from the instrument
- USB Export function now enabled
- Increased system memory stability
- Bug fixes
- Automated Workflow version checks to ensure compatibility
- Optimized CPU usage
- Fixed issue occasionally observed due to rapid back to back runs
- eProtein Discovery Instrument software / Firmware interface with improved logging
- Offline instrument (only for instruments installed to run in offline mode):
- Generic run can be created on the instrument without connection to the cloud software
- Constructs and cell-free blend will be differentiated with a standard naming convention and cannot be customized on screen
- Generic downselection of 30 highest expressing construct to the purification round
- USB only data export functionality enabled
- Generic run can be created on the instrument without connection to the cloud software
New workflow release: eProtein Discovery Screen Experiment 4.0.0
⚠ The workflow version will only be available once the instrument software has been updated to version 4.6.0. The following features will not be available retrospectively.
- Warnings are now displayed for measurements that may have been affected by droplet operation issues
- A Transfer Plate Guide can now be downloaded from the experiment's Progress tab
New workflow release: eProtein Discovery Screen Experiment 4.0.1
eProtein Discovery Screen Experiment 4.0.1 now replaces eProtein Discovery Screen Experiment 4.0.0 with the following updates:
- Fixed an issue where the "Top combinations for purified yield" table showed incorrect values
March 2024 releases
Instrument software release version 4.5.2
- Implementation of pin-based login and logout screen.
Password change and reset can be done on the eProtein Discovery Software by clicking on the user icon on the top right of the screen. Once changed, new pin will automatically apply to the instrument login screen
Other improvements:
-
- Better cartridge serial number detection and identification
- Cartridge actuation to increase droplet dispensing accuracy
New workflow version release: eProtein Discovery Screen Experiment 3.0.0
⚠ The new workflow version will only be available once the instrument software has been updated to version 4.5.2. The following features will not be available retrospectively.
- Added "Scale-Up Data" tab on the software for uploading scale-up data to completed experiments
- A PDF report is now included in the ZIP file on the instrument result screen and eProtein Discovery software
- Fixed bug that caused intermittent run failures
eProtein Discovery (cloud) software release
Users can now archive a protein, an experiment or a project generated on the eProtein Discovery Software (Retrieval of archived information is only available through Nuclera Technical Support).
- Software release notes are now available for access at any time, under the “help” menu.
- User feedback channel is now live, accessible through the “Help” menu.